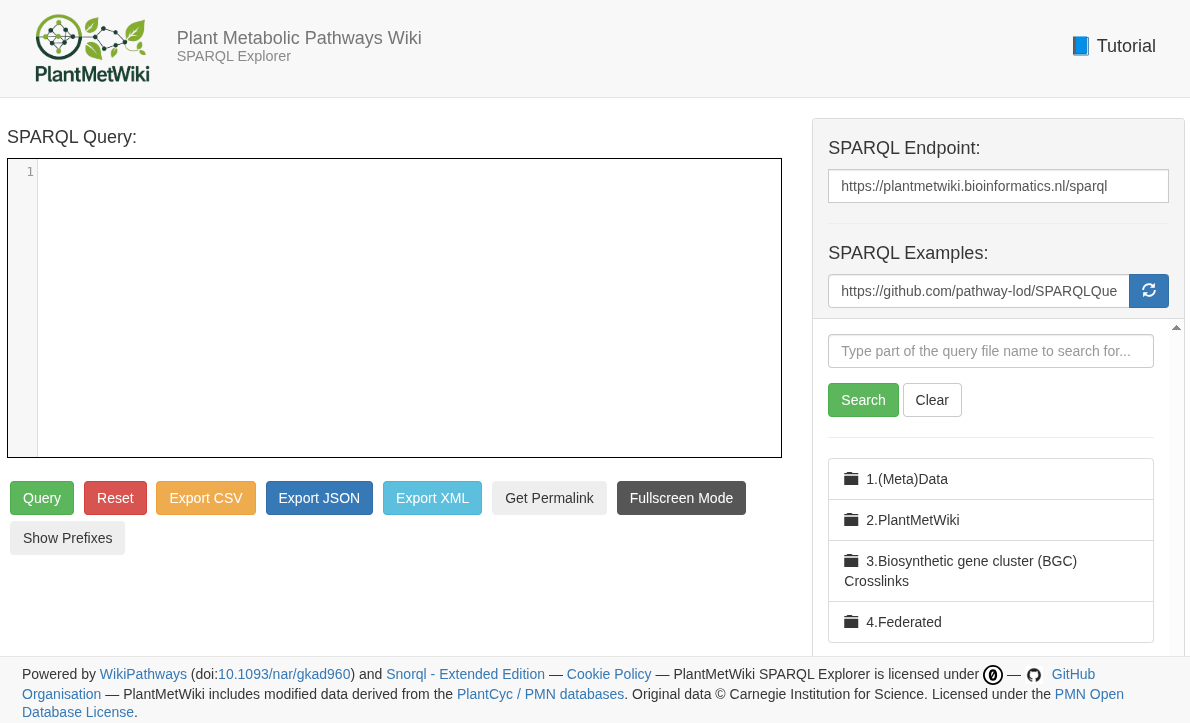
new blog post: "PlantMetWiki: a linked open data service for querying and analyzing plant pathway knowledge" https://chem-bla-ics.linkedchemistry.info/2026/01/05/plantmetwiki.html https://doi.org/10.59350/wyz5v-vts33
"I am happy that Elena and Denise pulled me into a collaboration to create an RDF-based knowledge graph about plant pathways."
tl;dr: https://plantmetwiki.bioinformatics.nl/
Replying to this post makes it show up in my blog.