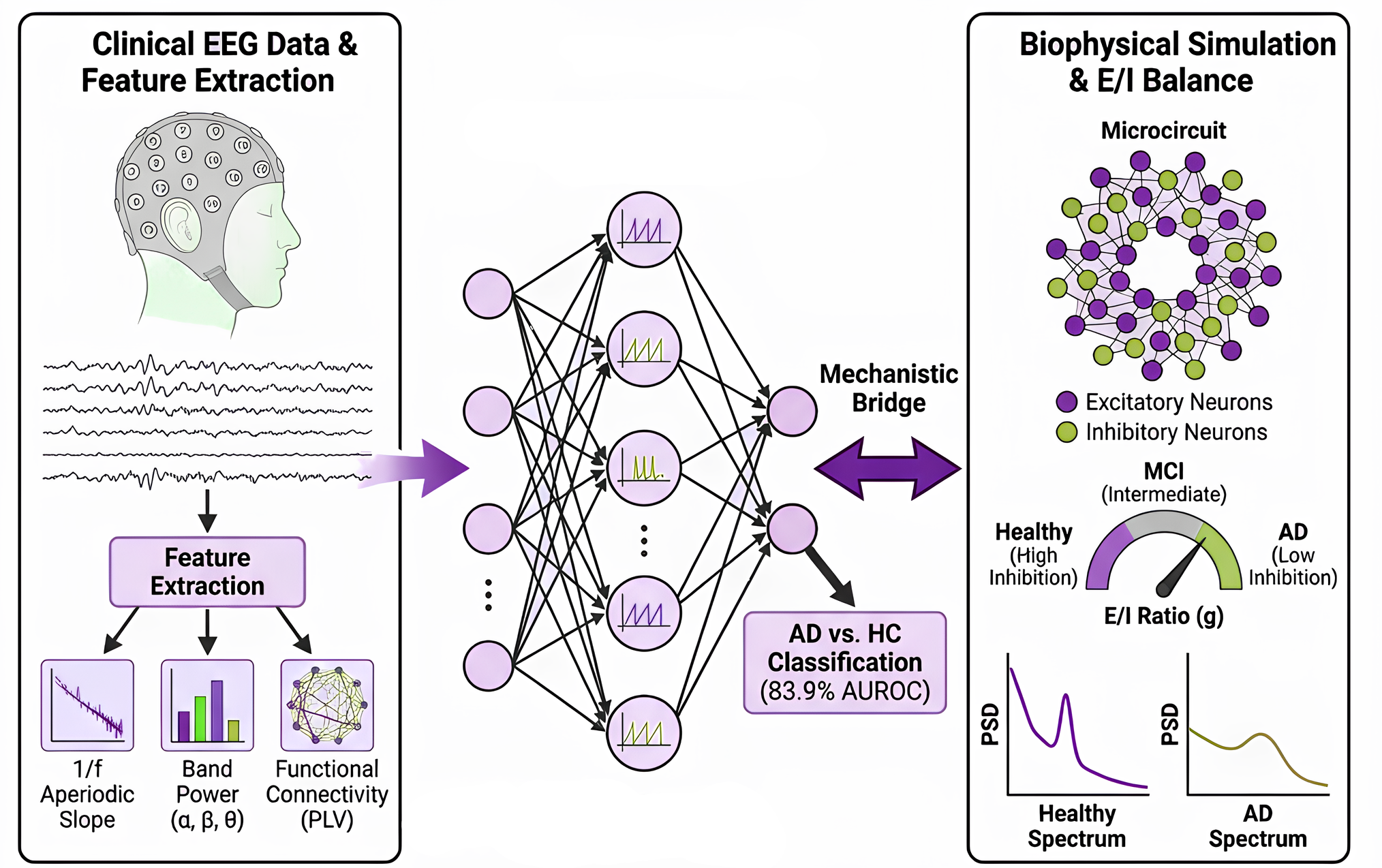
🧠Our New preprint:
We propose a “neuro-bridge” framework linking #spiking neural networks with biophysical brain simulations for Alzheimer’s detection from #EEG.
📉 1/f slope as key biomarker (E/I balance)
🔁 ML predictions ↔ circuit mechanisms #NeuroAI
https://arxiv.org/abs/2602.07010